The Mansfeld Lab
Our techniques
In vitro ubiquitination assays
To assay the activity of ubiquitin enzymes in defined conditions in vitro we express and purify all required components from E.coli, insect cells and human cell lines, respectively. The figure to the left illustrates an in vitro APC/C activity assay using ATP, methylated ubiquitin, GST-E1, His-UbcH10 and IRDye800-labelled securin as a substrate.
Note, methylated ubiquitin only supports mono-ubiquitination.
@Gabor Bakos

Fluorescent knock 'ins to visualize
endogenous proteins
To visualize endogenous proteins by live cell imaging we take advantage of pAAV or CRISPR/Cas9 assisted genome engineering. By targeting genes of fluorescent proteins site-specifc and in frame into the loci of genes of interest we determine protein dynamics during the cell cycle and the transition into quiescence. The movie to the right shows the dynamic beaviour of Cyclin A2 (blue), PCNA (red) and histone 3.1 (green) in proliferating retina pigment epithelial cells (RPE-1).
@Igor Gak

In vivo APC/C activity assays
To measure the activity of E3 ubiquitin ligases such as the APC/C in living cells we label their endogenous substrates (here Cyclin A2) with fluorescent proteins. By single cell time-lapse microscopy we monitor the degradation of such labelled substrates and extract flourescence changes over time as a readout of APC/C activity.
@Jörg Mansfeld
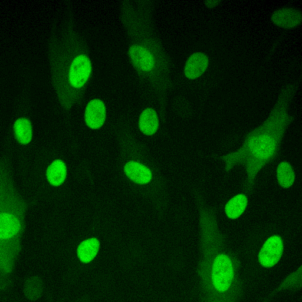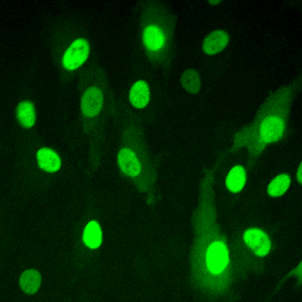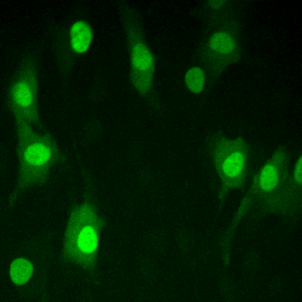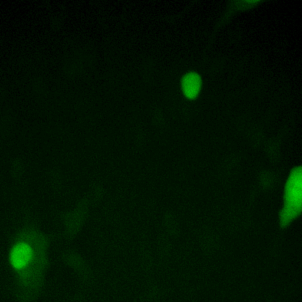
High resolution microscopy
@Magdalena Gonciarz

Immunofluorescent staining of endogenous proteins
Edit the text for the paragraph here
©Magdalena Gonciarz

Immunofluorescent staining of metaphase HeLa K cell.
Chromatin in blue (Hoechst) and tubulin in red.